In Saad et al. (Mol. Plant-Microbe Interact. 2019. 32:208–216), we studied the role of NifQ when Sinorhizobium fredii strain NGR234 enters in symbiosis with legume crops.
Rhizobia fix atmospheric nitrogen via the action of the nitrogenase enzyme. Normally, nitrogenase is only assembled when rhizobia are established inside plant cells of root nodules and are differentiating into functional bacteroids. Amongst the various enzymes required for nitrogenase assembly, NifQ was shown to donate molybdenum to the NifEN/NifH complexes.
Although NifQ was shown to be essential for nitrogenase assembly in free-living diazotrophs such as Klebsiella pneumoniae, many strains of rhizobia lack a functional nifQ gene. This suggests than in rhizobia NifQ is dispensable and its requirement might be dependent upon the host plant. In this respect, the promiscuous Sinorhizobium (Ensifer) fredii strain NGR234 is a model to assess the relative contribution of single gene products to many symbioses.
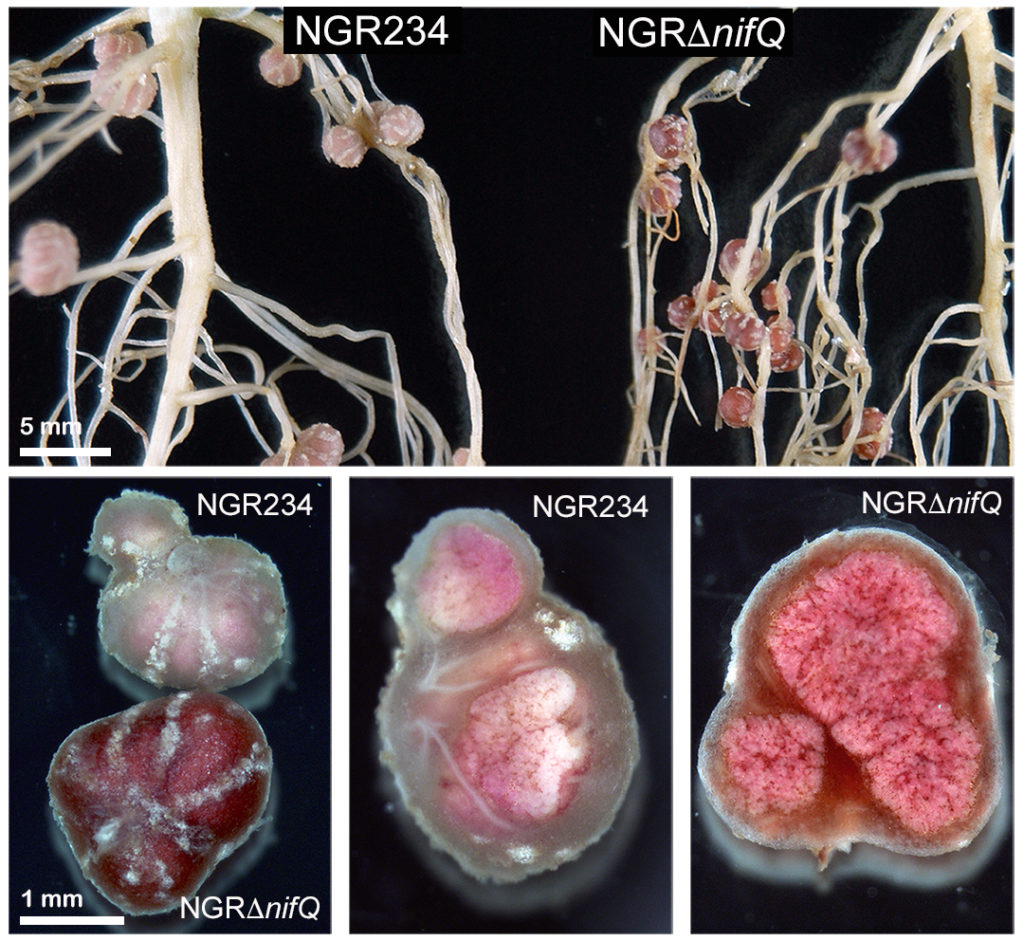
In this article, we describe how a deletion in nifQ of NGR234 (strain NGRΔnifQ) makes nodules of Vigna unguiculata (cowpea), V. radiata (mungbean), and Macroptilium atropurpureum (siratro) but not of the mimisoid tree Leucaena leucocephala, purple-red. This peculiar dark-nodule phenotype did not necessarily correlate with a decreased proficiency of NGRΔnifQ but coincided with a 20-fold or more accumulation inside cowpea nodules of coproporphyrin III and uroporphyrin III. Porphyrin accumulation was not restricted to plant cells infected with bacteroids but also extended to the nodule cortex. Nodule metal-homeostasis was altered but not sufficiently to prevent assembly and functioning of nitrogenase.
These results highlight how symbiotic proficiency remains a resilient process, with host plants apparently compensating for some deficiencies in rhizobia.